Motivation
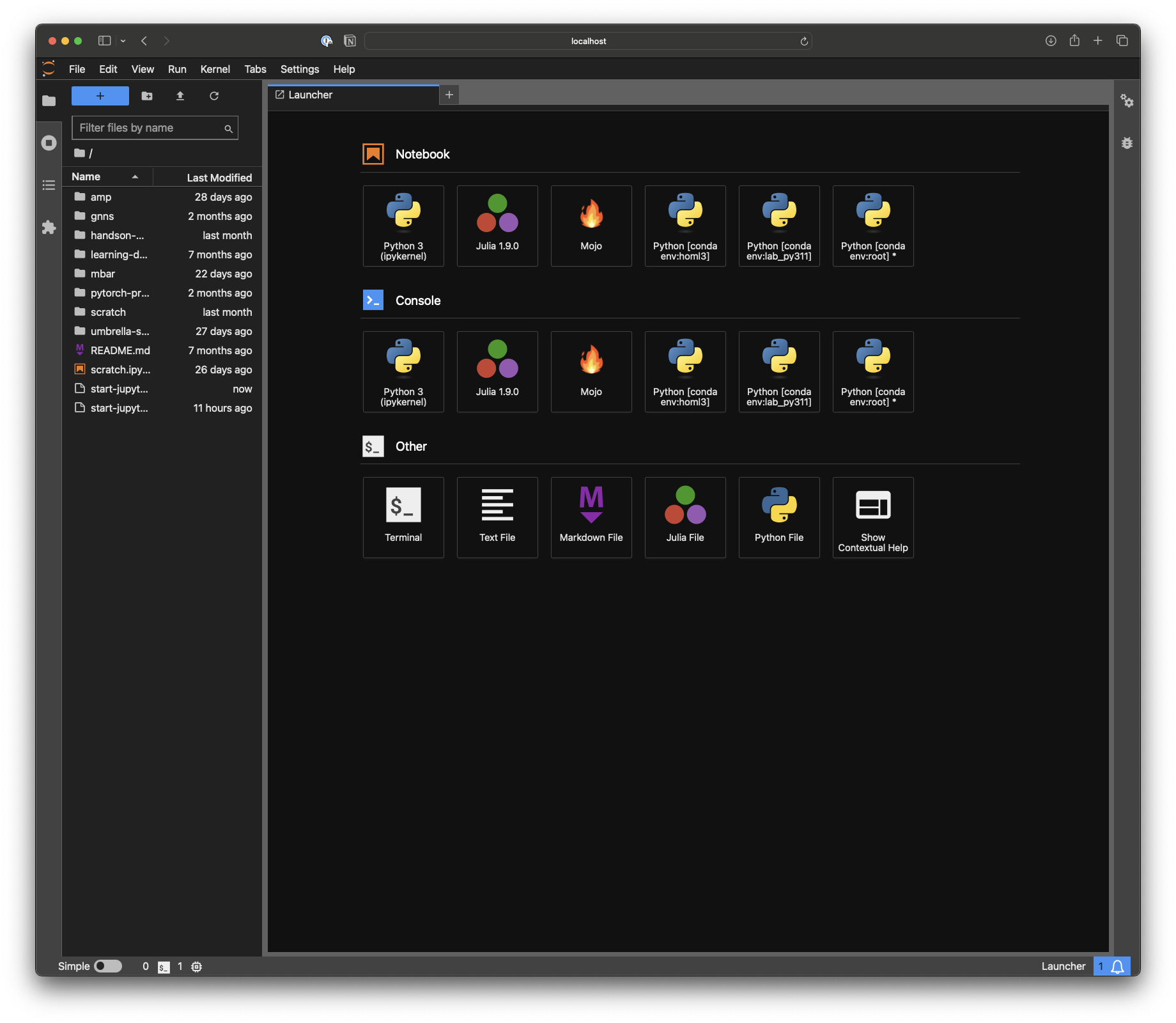
Whenever I want to quickly explore an idea, look at some data, generate a plot or would just like to run a simple calculation that is too complicated for Spotlight or Raycast, I like the option of just going to a local Jupyter lab instance on my Macbook or workstation. I have never been a fan of using Jupyter in VS Code or PyCharm. Aside from being more stable in my my experience, I love the simplicity of the browser for exploratory work. In the past, I found myself quite often accidentally closing the terminal window running the Jupyter instance and the connection dropped. I also found myself fighting quite a bit with the interplay of Miniconda environments and Jupyter. It happened quite often that I would start the Jupyter instance from the wrong environment or wanted to switch in the middle. Finally, starting Jupyter instances from many different folders of my computer led notebooks scattered all over my machines.
The First Part: nb_conda_kernels
There is the solution of manually adding every conda environment as a Jupyter kernel. I used to follow these instructions but often forgot to add a new environment created for a quick experiment. There is the much cleaner solution of nb_conda_kernels. Following the instructions on the developer's website, one simply installs this package into the base environment. Afterward, all conda environments that have the ipykernel package installed, will be visible from within Jupyter if the instance was started from base. For example, the following should work:
mamba activate base
mamba install nb_conda_kernels
mamba create -n lab_py311
mamba install -n lab_py311 python=3.11 ipykernel
jupyter lab
The commands assume you have mamba installed for faster installations.
The Second Part: nohup
One of my favorite UNIX command line tools is nohup (short for: no hang up). Preceding any command with nohup and following it with ampersand & causes the submitted process to be disowned and run in the background. I often do that for longer running jobs on cluster login nodes, e.g. copying files or rsync, or when I submit calculations or neural network trainings on my office workstation that will run overnight. The latter use case is especially important, when I submit the process via ssh. It has happened too often that I close the lid of my Macbook and am ready to call it a day, just to hear an almost immediate shutdown of the GPU fans one meter next to me. Coming back to the problem: on every computer I intent to use Jupyter, I create a folder called lab in my home directory. Within that folder, all my local projects will live and from there I start a single Jupyter instance using nohup:
#!/bin/zsh
cd $HOME/lab
echo "Starting Jupyter server at $(date)" >> start-jupyter-server.out
nohup jupyter lab --no-browser >> start-jupyter-server.out 2>&1 &
I typically request no browser window be opened and stderr be redirected to stdout. As long as my computer is running, I can go to http://localhost:8888/lab and work on a project or run a scratch notebook without the worry of accidentally closing a terminal window too much (Cmd+Q). Whenever I work on a new project, I simply create a new subfolder within lab. Finally, I added this script to my Login Items, so I never forget to run it after a reboot. I found that I had to change the default application for shell scripts to be iTerm or Terminal, otherwise Xcode would open upon login with an instance of the script instead.
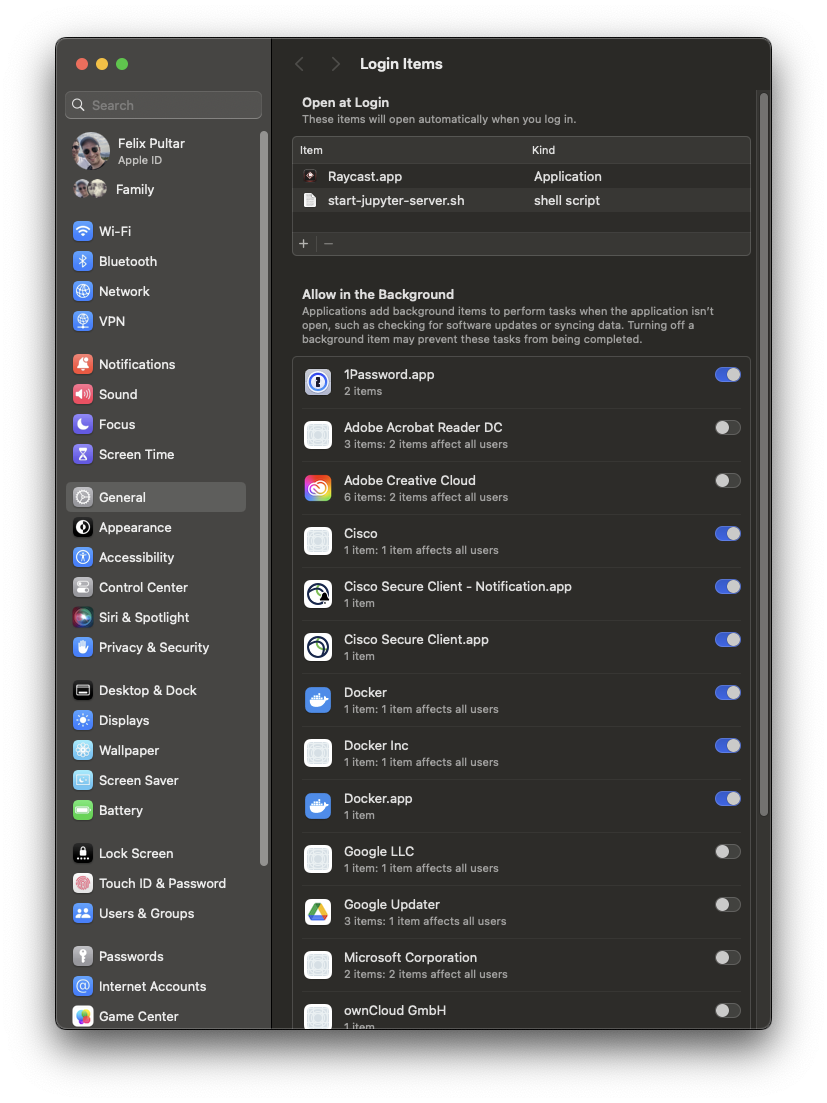
Conclusion
Following these steps, I can always spin up a new Jupyter notebook with an existing or new conda environment - it's that simple. mojo and julia are also installed on my system. I have another trick that extends this set-up in order to access an always running remote Jupyter instance, e.g. the one on my GPU workstation. I will share some details in a future post.